Polymerase chain reaction (PCR) is a powerful tool for detecting, diagnosing, and monitoring the presence of disease causing organisms in fish and shellfish.
It has many advantages over other traditional diagnostic methods such as culture-based procedures or serological tests.
The article described the details of Polymerase Chain Reaction principles, applications and procedure for aquatic animal disease diagnosis.
What is Polymerase Chain Reaction (PCR)
Polymerase Chain Reaction (PCR) is a laboratory technique used to amplify a specific DNA sequence, producing millions or billions of copies of the target DNA in a short period of time.
This process allows for the detection and analysis of small amounts of DNA, even from highly degraded or fragmented samples.
By repeatedly cycling through these steps, the amount of target DNA is exponentially increased, generating enough material for further analysis.
The process involves multiple cycles of heating and cooling, during which DNA is denatured, annealed with primers, and extended by a DNA polymerase enzyme.
This technique has numerous applications in various fields such as genetic research, medical diagnostics, forensic science, and environmental testing.
It has revolutionized the way scientists study DNA, allowing them to analyze even tiny amounts of genetic material quickly and accurately.
History of PCR
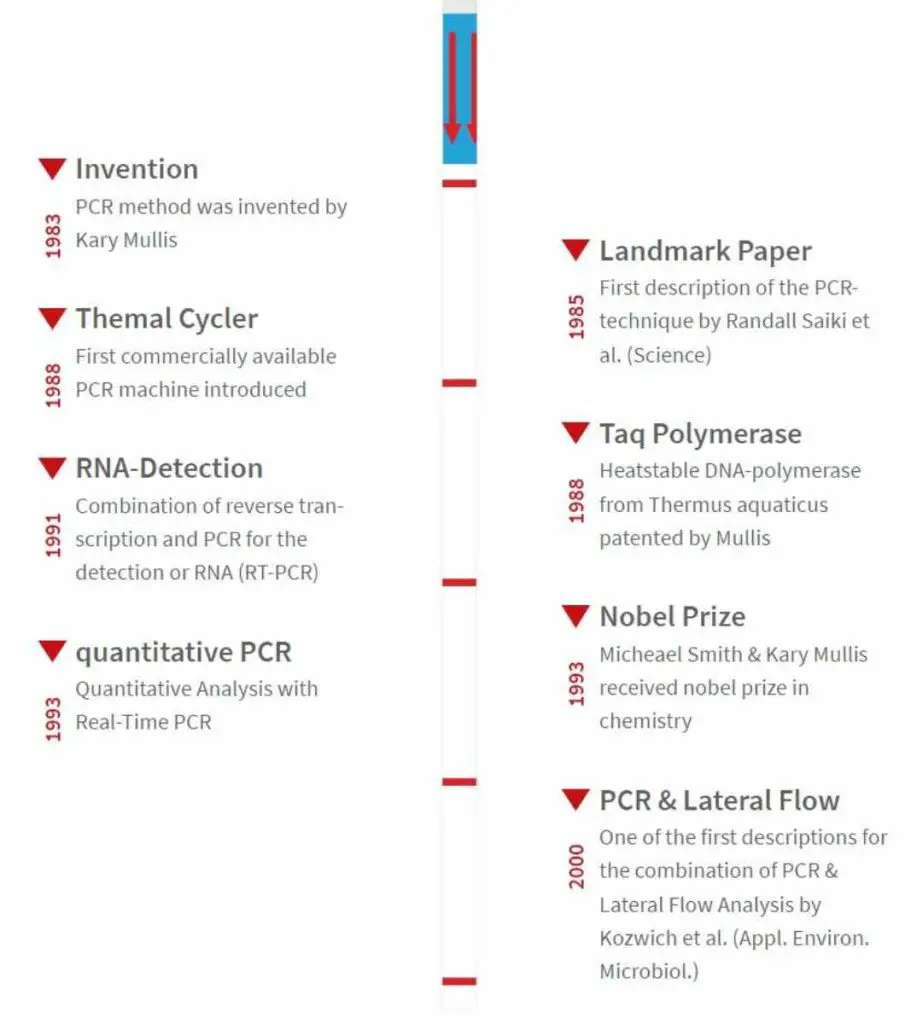
The Polymerase Chain Reaction (PCR) was first developed in the mid-1980s and has since revolutionized the field of molecular biology, genetics, and medicine.
PCR was first developed in 1983 by Kary Mullis, an American biochemist who was working for the Cetus Corporation at the time.
Mullis was trying to find a way to quickly and efficiently amplify DNA segments for use in genetic research.
His breakthrough came when he realized that he could use a heat-stable DNA polymerase enzyme to repeatedly copy a specific DNA sequence.
Initially, PCR was used for genetic research, allowing scientists to quickly and accurately amplify and analyze DNA sequences.
It was also used in forensic science, allowing for the analysis of small DNA samples found at crime scenes. The technique has since found a wide range of applications in medicine, agriculture, and biotechnology.
In the years following its development, PCR has undergone several improvements and modifications, including the development of real-time PCR, which allows for the quantification of DNA amplification in real-time, and multiplex PCR, which allows for the simultaneous amplification of multiple DNA segments.
PCR Applications | Roles of PCR in Disease Diagnosis
It offers several advantages over traditional diagnostic methods. Here are some of the roles of PCR in fish and shellfish disease diagnosis:
1. Identification of Pathogens
PCR can be used to identify specific pathogens (Bacteria, virus, fungus & parasite) that are causing diseases in fish and shellfish.
By designing specific primers for the target pathogen, PCR can amplify and detect even small amounts of pathogen DNA in the infected tissue or fluid samples.
2. Genetic Engineering
Polymerase Chain Reaction (PCR) is a powerful tool in genetic engineering, a field of biology that involves the manipulation of genes and genomes for practical purposes.
PCR allows for the amplification of specific DNA sequences, making it an essential technique in the creation of recombinant DNA molecules, gene cloning, and genetic modification.
PCR is used in genetic engineering to amplify specific DNA sequences, such as genes, promoters, and regulatory elements, which can then be inserted into other DNA molecules.
PCR-amplified DNA fragments can be easily inserted into plasmids, viruses, or other vectors, which can then be used to introduce the DNA fragment into a host cell or organism.
The method is also used to create mutations in DNA sequences, which can be useful in studying gene function or creating new genetic variants.
Site-directed mutagenesis, which involves the use of specific primers to introduce a desired mutation, is a common PCR-based technique used in genetic engineering.
3. Sanger Sequencing and NGS Applications
Polymerase Chain Reaction (PCR) is widely used in sequencing applications due to its ability to amplify specific DNA sequences from a complex mixture of genomic DNA.
PCR amplification is often used to generate DNA fragments for Sanger sequencing, a widely used method for determining the sequence of a DNA molecule.
PCR-based sequencing involves designing specific primers that flank the region of interest, and using these primers to amplify the region of interest using PCR.
The resulting PCR product can then be purified and subjected to Sanger sequencing, which involves using fluorescently-labeled dideoxynucleotides to terminate DNA synthesis at specific positions.
The resulting sequence data can be analyzed to determine the nucleotide sequence of the original DNA template.
In NGS, PCR amplification is used to generate millions of fragments, each of which is sequenced in parallel to generate massive amounts of sequence data.
This data can be used to assemble a complete genome or transcriptome, or to identify mutations, polymorphisms, and other genetic variations.
4. Forensic Science
PCR is used in forensic science to analyze DNA evidence in criminal investigations and identify suspects.
The process used to amplify specific DNA sequences from small amounts of biological material, such as blood, semen, or hair, which can then be used to generate a DNA profile or “genetic fingerprint” of the individual.
The resulting PCR products are then separated by size using gel electrophoresis, and the resulting band pattern is analyzed to generate a DNA profile.
This DNA profile can then be compared to DNA profiles from known individuals, such as suspects or victims, to determine if there is a match.
In addition, PCR-based DNA profiling has been used in paternity testing, immigration testing, and other applications where accurate individual identification is required.
5. Rapid Diagnosis
This rapid diagnosis method can produce results within a few hours. This allows for quick decision-making by physicians, farmers, veterinarians, and fisheries managers, which can prevent the spread of diseases and reduce economic losses.
6. Early Detection
PCR can detect pathogens at early stages of infection, before the onset of clinical signs, which is important for disease control and prevention.
Early detection allows for the implementation of appropriate management strategies, such as quarantine, treatment, or vaccination.
7. Environmental Monitoring
PCR can be used to monitor the presence of pathogens in the environment, such as in water or sediment samples.
This can provide early warning of potential disease outbreaks and help in the development of preventive measures.
PCR Types and Applications
There are several types of PCR that have been developed for different applications. The following are the types of PCR and their uses:
1. Standard PCR
Standard PCR is the most basic type of PCR, which amplifies a specific target DNA sequence.
It uses two primers that anneal to complementary regions of the target DNA sequence and DNA polymerase to synthesize new strands of DNA.
Standard PCR is used for DNA cloning, gene expression analysis, and mutation detection.
2. Reverse Transcription PCR (RT-PCR)
RT-PCR is used to amplify RNA sequences by first converting the RNA to complementary DNA (cDNA) using reverse transcriptase enzyme.
The cDNA is then amplified using standard PCR. RT-PCR is used for gene expression analysis, viral load measurement, and detecting RNA viruses.
3. Real-Time PCR (qPCR)
Real-time PCR is also known as quantitative PCR (qPCR) and is used to measure the amount of target DNA in a sample.
It uses fluorescent dyes or probes that bind to the amplified DNA during the reaction. Real-time PCR is used for gene expression analysis, viral load measurement, and pathogen detection.
4. Multiplex PCR
Multiplex PCR amplifies multiple DNA targets in a single reaction using multiple sets of primers that target different DNA sequences. It is used for detecting multiple pathogens, identifying gene mutations, and forensic DNA analysis.
Polymerase Chain Reaction (PCR) Principles
The PCR process is based on the principles of complementary base pairing, DNA polymerization, and thermal cycling.
The following are the key principles of the PCR process:
Complementary base pairing: The PCR process relies on the complementary base pairing of DNA molecules. The DNA molecule consists of four nitrogenous bases: adenine (A), thymine (T), cytosine (C), and guanine (G).
The base pairs are held together by hydrogen bonds, with A always pairing with T and C always pairing with G.
DNA polymerization: PCR uses a DNA polymerase enzyme, which can synthesize new DNA strands by adding nucleotides in a 5′ to 3′ direction.
The DNA polymerase enzyme is heat-stable and can withstand the high temperatures required for PCR.
Thermal cycling: PCR involves a series of heating and cooling cycles that are designed to denature the double-stranded DNA, anneal the primers, and extend the DNA.
Each cycle doubles the number of DNA molecules, resulting in exponential amplification of the target DNA sequence.
Primers: The PCR process requires the use of primers, which are short DNA sequences that are complementary to the target DNA sequence.
The primers bind to the complementary regions of the single-stranded DNA template during the annealing step, providing a starting point for DNA polymerase to extend the DNA sequence.
Optimization: The success of the PCR process depends on several factors, including primer design, DNA template quality and quantity, DNA polymerase enzyme selection, and the cycling conditions.
These factors must be optimized to ensure reliable and reproducible results.
PCR Machine and Its Parts
A PCR (Polymerase Chain Reaction) machine, also known as a thermal cycler, is a laboratory instrument used to amplify DNA. It consists of the following parts:
Thermal Block: This is the main part of the PCR machine that houses a block of metal or plastic with wells that hold the PCR tubes or plates containing the reaction mixture.
Heating and Cooling System: The thermal block is equipped with a heating and cooling system that can rapidly cycle the temperature of the reaction mixture to specific temperatures required for different steps of the PCR process.
Lid: The thermal block is covered by a lid that prevents contamination and evaporation of the reaction mixture during the PCR process.
Control Panel: The PCR machine has a control panel that allows the user to program and adjust the temperature settings and duration for each step of the PCR process.
Display Screen: The control panel includes a display screen that shows the current temperature and time of the PCR process.
Power Supply: The PCR machine is powered by an external power supply that provides the necessary electricity to run the machine.
Components of PCR
The process involves several components and chemicals, including:
DNA template: The DNA template is the sample that needs to be amplified.
Primers: Primers are short synthetic DNA sequences that are designed to bind to the specific regions of the template DNA that flanks the region of interest. The primers are critical for initiating the amplification process.
Taq polymerase: Taq polymerase is a heat-stable DNA polymerase enzyme that catalyzes the synthesis of new DNA strands from the primers. It is used because it can withstand the high temperatures used during the PCR process.
Deoxyribonucleotide triphosphates (dNTPs): dNTPs are the building blocks of DNA. They are the four nucleotides (adenine, guanine, cytosine, and thymine) that are incorporated into the newly synthesized DNA strands by Taq polymerase during the PCR process.
Buffer solution: The buffer solution contains various salts and pH stabilizers that provide optimal conditions for the Taq polymerase to function.
Magnesium ions (Mg2+): Magnesium ions are essential cofactors for Taq polymerase activity.
Polymerase Chain Reaction Steps | PCR Procedure or Process
The Polymerase Chain Reaction (PCR) process consists of three main steps: denaturation, annealing, and extension.
The following is a step-by-step description of the PCR process:
1. Denaturation
The first step in the PCR process is to denature the double-stranded DNA template into two separate single strands.
This is achieved by heating the DNA to a high temperature (typically 94-98°C) for a short period of time (usually 30 seconds to 1 minute).
2. Annealing
After denaturation, the temperature is lowered to allow the primers to anneal to the complementary regions of the single-stranded DNA template.
The primers are short, synthetic DNA sequences that are designed to bind specifically to the target DNA sequence at the ends of the region to be amplified.
The temperature for annealing is typically 50-65°C, and the length of time depends on the specific primers used.
3. Extension
Once the primers have annealed, the temperature is raised again to allow the DNA polymerase enzyme to extend the primers and synthesize a new complementary DNA strand from each single-stranded template.
The DNA polymerase enzyme adds nucleotides to the 3′ end of the primers, building a new strand of DNA in the 5′ to 3′ direction. The optimal temperature for the DNA polymerase enzyme to work is typically around 72°C.
The length of time for extension depends on the length of the DNA template and the rate of polymerization of the enzyme.
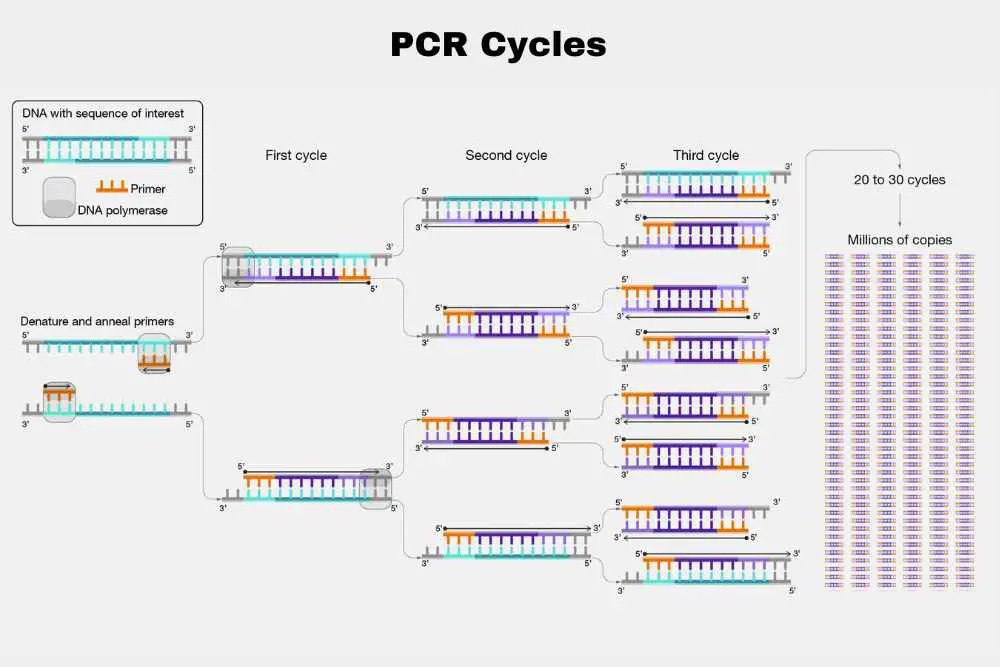
After the first cycle of denaturation, annealing, and extension, the process is repeated for a set number of cycles, usually 20-40 times.
Each cycle doubles the amount of DNA in the sample, resulting in an exponential increase in the number of copies of the target DNA sequence.
The end result of the PCR process is the amplification of a specific DNA sequence, which can then be analyzed by various methods, such as gel electrophoresis, sequencing, or hybridization.
Polymerase Chain Reaction (PCR) Limitations
Despite its numerous benefits, PCR (Polymerase Chain Reaction) has several limitations that researchers should be aware of:
False positives: Contamination of the PCR reaction with DNA from other sources can lead to false positives, which can be difficult to identify and eliminate.
Limited sample size: The amount of DNA in a sample is limited, and PCR may not be able to amplify a sufficient amount of DNA for analysis.
Amplification bias: PCR may not amplify all DNA sequences equally, resulting in an amplification bias that can affect the accuracy of the results.
Specificity issues: PCR may amplify non-specific DNA sequences, leading to false-positive results or interfering with the detection of the target DNA sequence.
Fragment length limitations: PCR has a limit to the length of DNA fragments that can be amplified. The length of the fragment depends on the specific PCR conditions and the enzyme used.
Enzyme limitations: PCR requires the use of a specific type of enzyme, which may not be suitable for certain applications.
Cost: PCR can be expensive, especially when using high-quality reagents and equipment.
Time-consuming: PCR is a time-consuming technique that requires several hours or days to complete, depending on the specific protocol.
PCR Machines
1. Biometra TAdvanced Thermal Cycler Series (Analytik Jena)
The Biometra TAdvanced Thermal Cycler Series features a block that can hold 48 or 96 PCR tubes, allowing for high-throughput PCR applications.
It also has a large touch screen display for easy programming and monitoring of PCR cycles.
The instrument uses Peltier elements for rapid heating and cooling, and has gradient capability for optimizing PCR conditions.
Advantages: The high-throughput capacity of the Biometra TAdvanced Thermal Cycler Series makes it suitable for applications such as genotyping, gene expression analysis, and cloning.
The gradient capability allows for optimization of PCR conditions, increasing the accuracy and reproducibility of results.
Limitations: The Biometra TAdvanced Thermal Cycler Series can be relatively expensive, and its high-throughput capacity may not be necessary for all applications.
2. MiniAmp™ Plus Thermal Cycler (ThermoFisher Scientific):
The MiniAmp™ Plus Thermal Cycler is a compact instrument that can hold up to 16 PCR tubes.
It has a simple user interface with intuitive controls and an LED display. The instrument uses Peltier elements for rapid heating and cooling.
Advantages: The MiniAmp™ Plus Thermal Cycler is compact and easy to use, making it suitable for small laboratories or fieldwork. Its affordability makes it an attractive option for labs with limited budgets.
Limitations: The MiniAmp™ Plus Thermal Cycler’s limited capacity may not be suitable for high-throughput applications. It also lacks gradient capability, which may limit its use in optimizing PCR conditions.
3. Biometra TAdvanced Thermal Cycler Series (Analytik Jena)
The Biometra TAdvanced Thermal Cycler Series is a high-performance PCR machine that can accommodate various sample formats and reaction volumes.
It has a large color touch screen interface for easy programming and monitoring of the PCR reactions.
It also has a heated lid with adjustable pressure to prevent evaporation and ensure uniform heating.
Uses: This thermal cycler is suitable for a wide range of PCR applications, including genotyping, gene expression analysis, and DNA sequencing. Its flexible block format allows for the simultaneous amplification of multiple samples.
Limitations: The Biometra TAdvanced Thermal Cycler Series is a high-end instrument with a relatively high price point. It may not be affordable for smaller research labs or budget-limited projects.
4. PCR Thermal Cyclers (Esco)
Esco’s PCR Thermal Cyclers are designed to provide accurate and consistent temperature control for PCR reactions. They have a variety of block formats to accommodate different sample sizes and types.
The user interface is intuitive, with a large color touch screen and easy-to-use software.
Uses: Esco’s PCR Thermal Cyclers are suitable for a wide range of PCR applications, including genotyping, gene expression analysis, and DNA sequencing. They can also be used for in situ PCR and in vitro transcription applications.
Limitations: Esco’s PCR Thermal Cyclers are relatively expensive compared to some other models on the market. They may not be suitable for smaller research labs or budget-limited projects.
5. MiniAmp™ Plus Thermal Cycler (ThermoFisher Scientific)
The MiniAmp™ Plus Thermal Cycler is a compact and portable PCR machine that can accommodate up to 24 samples in a single run.
It has a simple user interface with a small LCD display and intuitive navigation buttons.
Uses: The MiniAmp™ Plus Thermal Cycler is ideal for small-scale PCR applications, such as routine genotyping and screening of gene mutations. Its small size and portability make it suitable for fieldwork and point-of-care diagnostics.
Limitations: The MiniAmp™ Plus Thermal Cycler has a limited capacity of up to 24 samples per run. It may not be suitable for larger-scale PCR projects.
6. GET-S SERIES THERMAL CYCLER (Bio-gener)
The GET-S SERIES THERMAL CYCLER is a versatile PCR machine with a modular design that allows for customization based on specific research needs.
It has a large color touch screen interface with user-friendly software for programming and monitoring PCR reactions.
Uses: The GET-S SERIES THERMAL CYCLER is suitable for a wide range of PCR applications, including genotyping, gene expression analysis, and DNA sequencing. Its modular design allows for easy customization for specific research needs.
Limitations: The GET-S SERIES THERMAL CYCLER is relatively expensive compared to some other models on the market. It may not be suitable for smaller research labs or budget-limited projects.
FAQs
What is a PCR machine called?
A PCR machine is typically called a thermal cycler or PCR cycler. This instrument is designed to precisely control the temperature of the PCR reaction, allowing for the repeated cycles of heating and cooling necessary for DNA amplification to occur.
Why is PCR cycle repeated 30 times?
PCR cycles are repeated typically around 30 times to ensure that there is enough amplification of the target DNA sequence. Each cycle doubles the amount of the target DNA sequence, and after 30 cycles, there can be up to a billion copies of the original DNA segment.
Does PCR require ATP?
Yes, PCR requires ATP (adenosine triphosphate) as a source of energy for the DNA polymerase enzyme that synthesizes new DNA strands during the reaction.
What is the size of PCR?
The size of a PCR product can vary depending on the specific target sequence being amplified and the length of the primers used in the reaction. However, PCR can typically amplify DNA fragments ranging from a few hundred base pairs to several thousand base pairs in length.
What length of DNA is used in PCR?
The length of the DNA segment used in PCR depends on the specific application and the target DNA sequence being amplified. PCR can be used to amplify a specific region of a longer DNA molecule, such as a gene or a regulatory element.
How to calculate PCR DNA?
To calculate the amount of DNA generated by PCR, you can use the following formula:
DNA (in ng) = (concentration of PCR product in ng/μL) x (total volume of PCR product in μL)
What is the mixture of PCR reaction?
The mixture of a typical PCR reaction includes the target DNA template, DNA primers, DNA polymerase enzyme, deoxynucleoside triphosphates (dNTPs), buffer solution, and water.
Is PCR anaerobic?
No, PCR does not necessarily need to be anaerobic, but it is typically performed under strict temperature control to prevent the degradation of the DNA template and to optimize the efficiency of the reaction.
References
- Erlich, H. A. (1989). Polymerase chain reaction. Journal of clinical immunology, 9, 437-447.
- Bartlett, J. M., & Stirling, D. (2003). A short history of the polymerase chain reaction. PCR protocols, 3-6.
- Kubista, M., Andrade, J. M., Bengtsson, M., Forootan, A., Jonák, J., Lind, K., & Zoric, N. (2006). The real-time polymerase chain reaction. Molecular aspects of medicine, 27(2-3), 95-125.
- Joshi, M., & Deshpande, J. D. (2010). Polymerase chain reaction: methods, principles and application. International Journal of Biomedical Research, 2(1), 81-97.
- Tattiyapong, P., Sirikanchana, K., & Surachetpong, W. (2018). Development and validation of a reverse transcription quantitative polymerase chain reaction for tilapia lake virus detection in clinical samples and experimentally challenged fish. Journal of Fish Diseases, 41(2), 255-261.
- Brown, L. L., Iwama, G. K., Evelyn, T. P. T., Nelson, W. S., & Levine, R. P. (1994). Use of the polymerase chain reaction (PCR) to detect DNA from Renibacterium salmoninarum within individual salmonid eggs. Diseases of aquatic organisms, 18(3), 165-171.
- Altinok, I., Grizzle, J. M., & Liu, Z. (2001). Detection of Yersinia ruckeri in rainbow trout blood by use of the polymerase chain reaction. Diseases of Aquatic Organisms, 44(1), 29-34.
- Gilad, O., Yun, S., Andree, K. B., Adkison, M. A., Zlotkin, A., Bercovier, H., … & Hedrick, R. P. (2002). Initial characteristics of koi herpesvirus and development of a polymerase chain reaction assay to detect the virus in koi, Cyprinus carpio koi. Diseases of aquatic organisms, 48(2), 101-108.
- Gustafson, C. E., Thomas, C. J., & Trust, T. (1992). Detection of Aeromonas salmonicida from fish by using polymerase chain reaction amplification of the virulence surface array protein gene. Applied and Environmental Microbiology, 58(12), 3816-3825.
- Palenzuela, O., Trobridge, G., & Bartholomew, J. L. (1999). Development of a polymerase chain reaction diagnostic assay for Ceratomyxa shasta, a myxosporean parasite of salmonid fish. Diseases of aquatic organisms, 36(1), 45-51.
- Bilodeau, A. L., Waldbieser, G. C., Terhune, J. S., Wise, D. J., & Wolters, W. R. (2003). A real-time polymerase chain reaction assay of the bacterium Edwardsiella ictaluri in channel catfish. Journal of Aquatic Animal Health, 15(1), 80-86.
- Valverde, E. J., Cano, I., Labella, A., Borrego, J. J., & Castro, D. (2016). Application of a new real-time polymerase chain reaction assay for surveillance studies of lymphocystis disease virus in farmed gilthead seabream. BMC veterinary research, 12(1), 1-8.
